Abstract
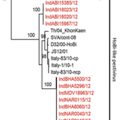
HoBi-like pestiviruses have been sporadically reported from naturally infected cattle in South America, Asia and Europe. While the closely related bovine viral diarrhoea virus 1 (BVDV-1) and BVDV-2 have been reported from cattle in India, the prevalence and diversity of HoBi-like viruses have not yet been studied. Here we report the genetic diversity and molecular characteristics of HoBi-like viruses, through systematic surveillance in cattle (n = 1049) from 21 dairy farms across India during 2012–2013. On the basis of real-time RT-PCR, virus isolation and nucleotide sequencing results, of the 20 pestivirus positive cattle, HoBi-like viruses were identified in 19 cattle from four farms in three states and BVDV-1b in one cattle. Phylogenetic analysis of 5′-UTR and Npro region identified the circulation of two lineages of HoBi-like viruses in India, that were distinct to those circulating globally, highlighting the independent evolution of at least three lineages of HoBi-like viruses globally. Antigenic differences were also evident between the two Indian lineages. In addition to revealing that HoBi-like virus may be more widespread in Indian cattle than previously reported, this study shows greater genetic divergence of HoBi-like viruses indicating a need for continued pestivirus surveillance in cattle.